| 利用NCBI当中的CD | 您所在的位置:网站首页 › ncbi数据库检索工具 › 利用NCBI当中的CD |
利用NCBI当中的CD
|
1.CDD入口:https://www.ncbi.nlm.nih.gov/cdd/,该工具为NCBI提供的,NCBI主页选项口选择进入Conserved Domain然后点击Search。 然后选择CD-search(单个基因),或者Batch CD-Search(多个基因)。
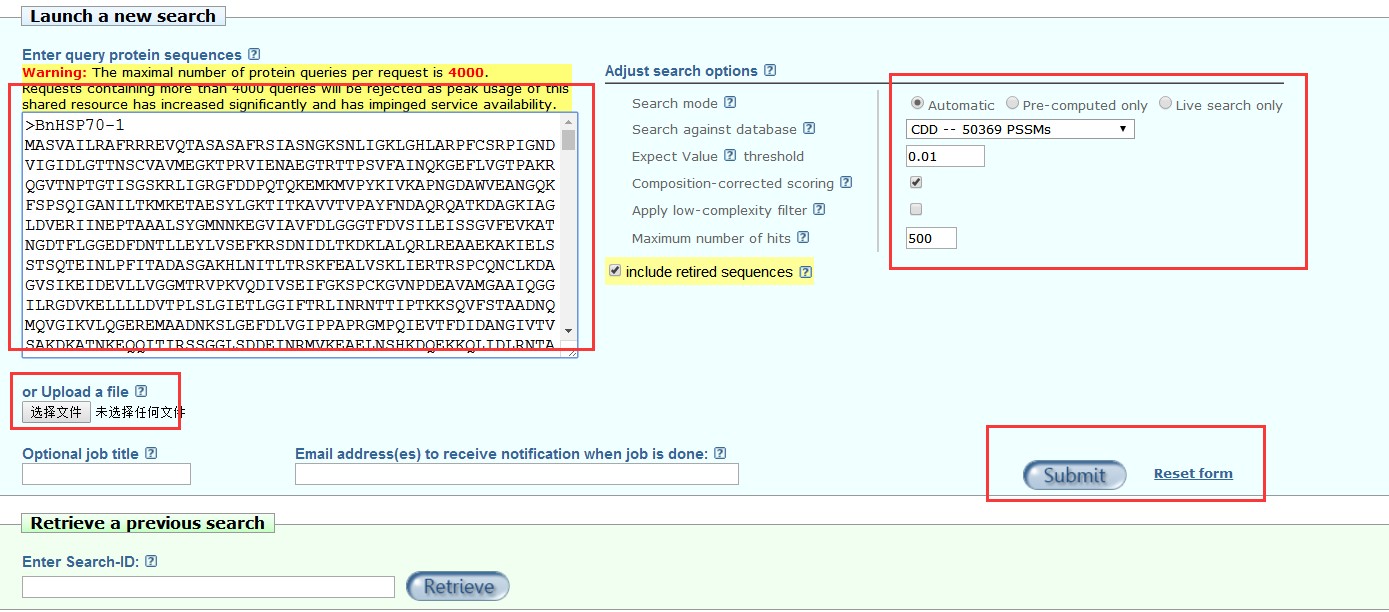
2.预测单条序列的保守结构域,点击CD-search进入: 粘贴基因的蛋白序列,或者是核酸序列都可以,注意为fasta序列,如果你知道基因的GI或Accession号,也可以直接数据这些ID就可以查询,右方OPTIONS中选择要搜索的数据库,Expect Value等,或者使用默认设置,然后按“提交”按钮。就可以搜索了;
3.结果查看,默认为concise 简洁显示,可以选择full result显示全部内容:
4.批量提交序列搜索保守结构域,Batch CD-Search,如下:: 可选择直接粘贴多序列的fasta格式文件,或者提交fasta格式的文件,然后选择数据库,如果数据较大搜索时间很久,可填写email,搜索完成之后会发邮寄,最后点击submit提交就可以搜索了:
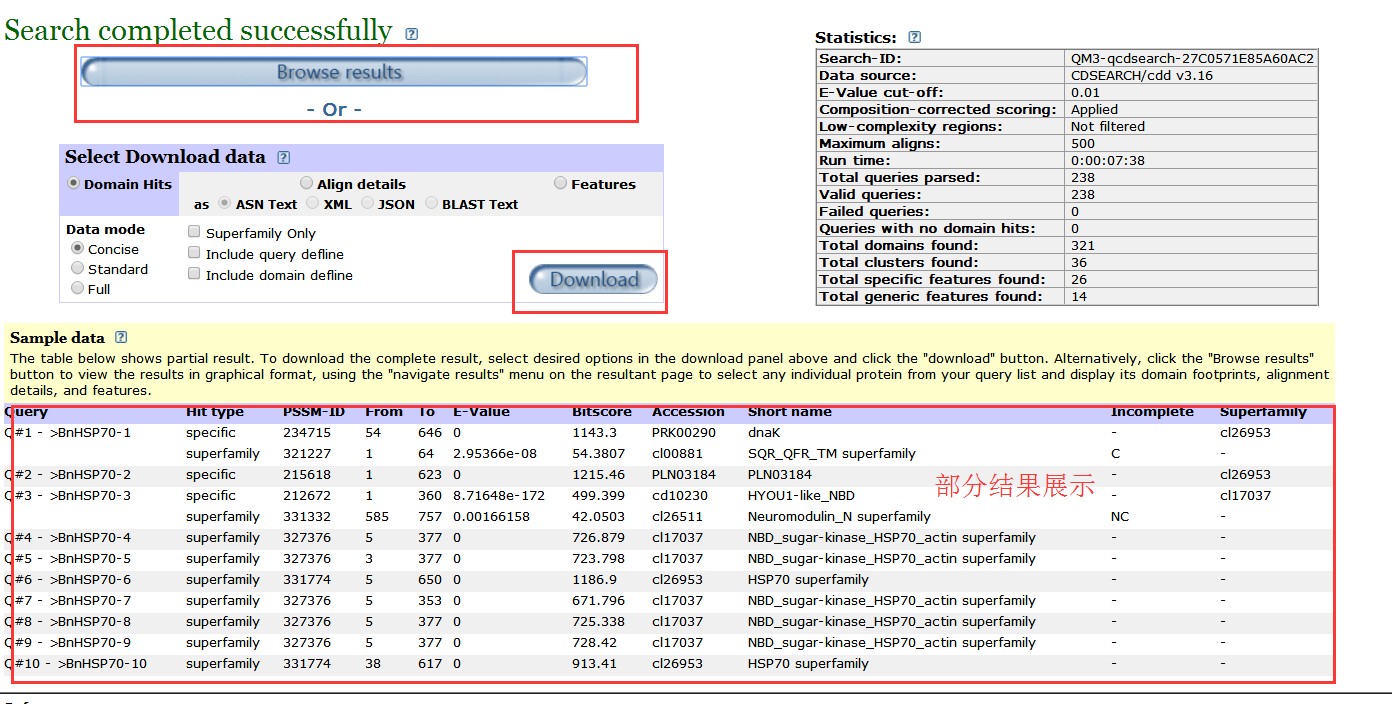
5.批量搜索结果展示: 批量搜索结果如下,只展示部分结果,详细结果可点击download进行下载(记得勾选full,下载全部结构域信息),或者点击Browse results 进行详细的可视化浏览;
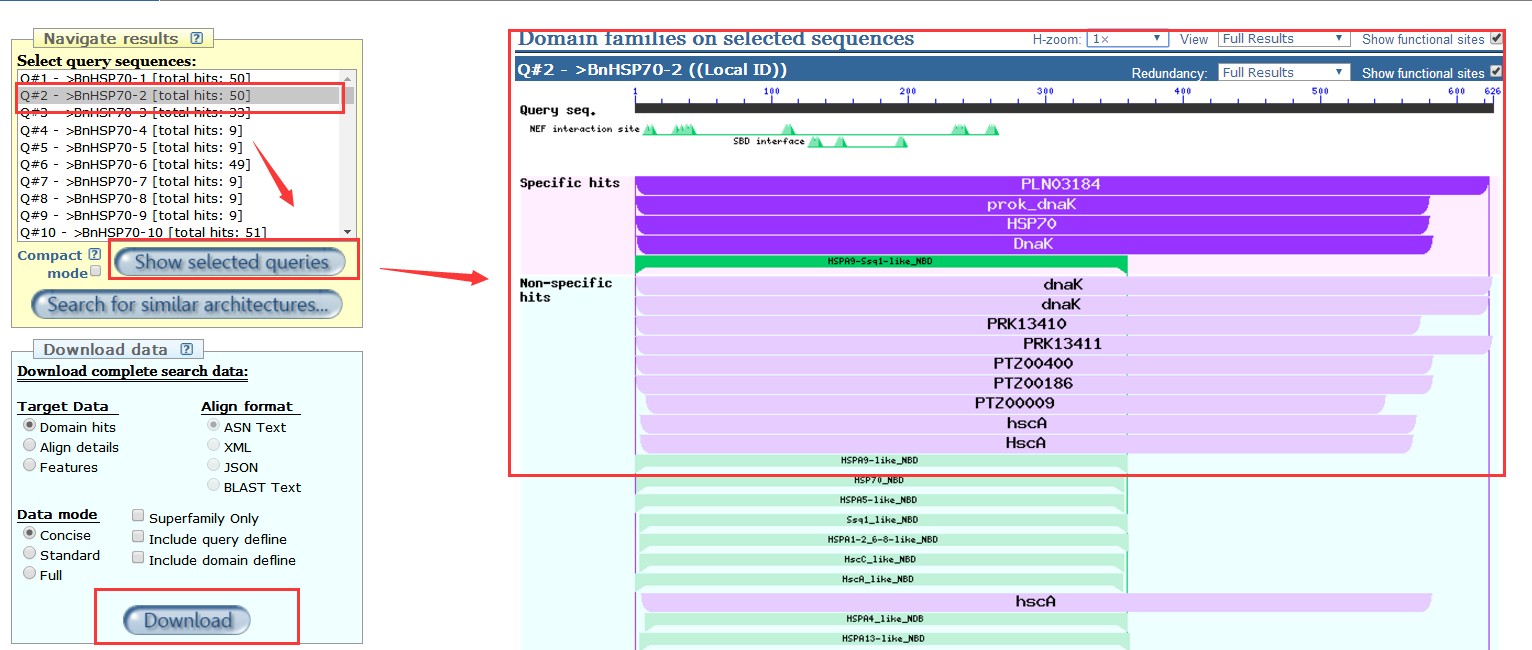
表格结果说明如下: Query输入的序列IDHit typeCD-Search results can include hit types that represent various confidence levels (specific hits, non-specific hits) and domain model scope (superfamilies, multi-domains). They can be seen in both the Concise display and Full display, except for non-specific hits, which are shown only in the Full Display.PSSM-IDA PSSM ID is the unique identifier for a domain model's position-specific scoring matrix (PSSM). From..ToThe range of amino acids in the query protein sequence to which the domain model aligns. (Note: If the alignment found by RPS-BLAST omitted more than 20% of the CD's extent at either the n- or c-terminus or both, the partial nature of the hit is indicated in the "Incomplete" column of the hit table. Partial hits can also be spotted in the graphical display as domain model cartoons with jagged edges (illustrated example).)E-valueThe expect value, or E-value, indicates the statistical significance of the hit as the likelihood the hit was found by chance. Bit Score比对得分AccessionThe accession number of the hit, which can either be a domain model or a superfamily cluster. (If the hit is a domain model, then the accession number (cl*) of the superfamily cluster to which it belongs is listed in the "Superfamily" column of the output file.)Short nameThe short name of a conserved domain, which concisely defines the domain. For example, "Voltage gated ClC" is the short title of the NCBI-curated conserved domain model for the voltage gated chloride channel (cd00400).IncompleteIf the hit to a conserved domain is partial (i.e., if the alignment found by RPS-BLAST omitted more than 20% of the CD's extent at either the n- or c-terminus or both), this column will be populated with one of the following values: N: incomplete at the N-terminus C: incomplete at the C-terminus NC: incomplete at both the N-terminus and C-terminusIf the hit to a conserved domain is complete, then this column will be populated with a dash (-).(Note: Partial hits can also be spotted in the graphical display as domain model cartoons with jagged edges (illustrated example).)SuperfamilyThis column is populated only for domain models that are specific or non-specific hits, and it lists the accession number of the superfamily to which the domain model belongs.(If the hit is to a superfamily itself, then this column is simply populated with a dash because the superfamily accession is already listed in the preceding "Accession" column.)6.批量搜索结果,可视化浏览界面: 可以选中,然后浏览,下载等:
 Marchler-Bauer A et al. (2017), "CDD/SPARCLE: functional classification of proteins via subfamily domain architectures.", Nucleic Acids Res.45(D)200-3. Marchler-Bauer A et al. (2017), "CDD/SPARCLE: functional classification of proteins via subfamily domain architectures.", Nucleic Acids Res.45(D)200-3. Marchler-Bauer A et al. (2015), "CDD: NCBI's conserved domain database.", Nucleic Acids Res.43(D)222-6. Marchler-Bauer A et al. (2015), "CDD: NCBI's conserved domain database.", Nucleic Acids Res.43(D)222-6. Marchler-Bauer A et al. (2011), "CDD: a Conserved Domain Database for the functional annotation of proteins.", Nucleic Acids Res.39(D)225-9. Marchler-Bauer A et al. (2011), "CDD: a Conserved Domain Database for the functional annotation of proteins.", Nucleic Acids Res.39(D)225-9. Marchler-Bauer A, Bryant SH (2004), "CD-Search: protein domain annotations on the fly.", Nucleic Acids Res.32(W)327-331.
更多基因家族分析内容,更多生物信息课程: Marchler-Bauer A, Bryant SH (2004), "CD-Search: protein domain annotations on the fly.", Nucleic Acids Res.32(W)327-331.
更多基因家族分析内容,更多生物信息课程:1. 文章越来越难发?是你没发现新思路,基因家族分析发2-4分文章简单快速,学习链接:基因家族分析实操课程、基因家族文献思路解读 2. 转录组数据理解不深入?图表看不懂?点击链接学习深入解读数据结果文件,学习链接:转录组(有参)结果解读;转录组(无参)结果解读 3. 转录组数据深入挖掘技能-WGCNA,提升你的文章档次,学习链接:WGCNA-加权基因共表达网络分析 4. 转录组数据怎么挖掘?学习链接:转录组标准分析后的数据挖掘、转录组文献解读 5. 微生物16S/ITS/18S分析原理及结果解读、OTU网络图绘制、cytoscape与网络图绘制课程 6. 生物信息入门到精通必修基础课:linux系统使用、biolinux搭建生物信息分析环境、linux命令处理生物大数据、perl入门到精通、perl语言高级、R语言画图、R语言快速入门与提高 7. 医学相关数据挖掘课程,不用做实验也能发文章:TCGA-差异基因分析、GEO芯片数据挖掘、 GEO芯片数据不同平台标准化 、GSEA富集分析课程、TCGA临床数据生存分析、TCGA-转录因子分析、TCGA-ceRNA调控网络分析 8.其他,二代测序转录组数据自主分析、NCBI数据上传、二代fastq测序数据解读、 9.更多课程可点击:组学大讲堂视频课程 |
【本文地址】
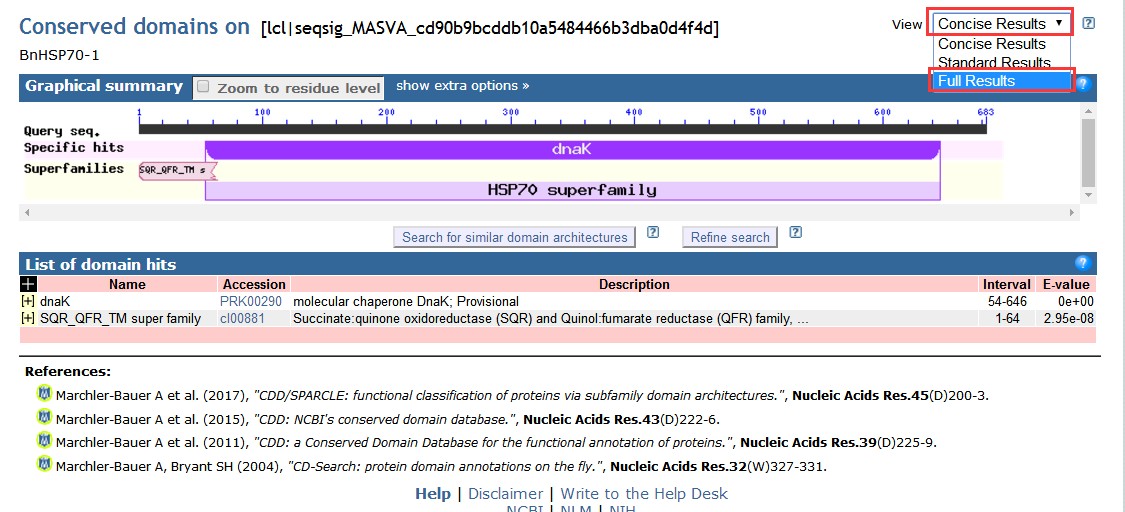 搜索匹配结果,有特定匹配(specific hits),非特定匹配(non-specific hits),这些匹配所属的超家族(superfamily),我们发现这个基因特定匹配在dnak,属于HSP70超级家族;
搜索匹配结果,有特定匹配(specific hits),非特定匹配(non-specific hits),这些匹配所属的超家族(superfamily),我们发现这个基因特定匹配在dnak,属于HSP70超级家族;