| Microorganisms | 您所在的位置:网站首页 › 天津卫视网络直播平台 › Microorganisms |
Microorganisms
|
Journals
Active Journals
Find a Journal
Proceedings Series
Topics
Information
For Authors
For Reviewers
For Editors
For Librarians
For Publishers
For Societies
For Conference Organizers
Open Access Policy
Institutional Open Access Program
Special Issues Guidelines
Editorial Process
Research and Publication Ethics
Article Processing Charges
Awards
Testimonials
Author Services
Initiatives
Sciforum
MDPI Books
Preprints.org
Scilit
SciProfiles
Encyclopedia
JAMS
Proceedings Series
About
Overview
Contact
Careers
News
Press
Blog
Sign In / Sign Up
Notice
clear
Notice
You are accessing a machine-readable page. In order to be human-readable, please install an RSS reader. Continue Cancel clearAll articles published by MDPI are made immediately available worldwide under an open access license. No special permission is required to reuse all or part of the article published by MDPI, including figures and tables. For articles published under an open access Creative Common CC BY license, any part of the article may be reused without permission provided that the original article is clearly cited. For more information, please refer to https://www.mdpi.com/openaccess. Feature papers represent the most advanced research with significant potential for high impact in the field. A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications. Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the reviewers. Editor’s Choice articles are based on recommendations by the scientific editors of MDPI journals from around the world. Editors select a small number of articles recently published in the journal that they believe will be particularly interesting to readers, or important in the respective research area. The aim is to provide a snapshot of some of the most exciting work published in the various research areas of the journal. Original Submission Date Received: .  Journals
Active Journals
Find a Journal
Proceedings Series
Topics
Information
For Authors
For Reviewers
For Editors
For Librarians
For Publishers
For Societies
For Conference Organizers
Open Access Policy
Institutional Open Access Program
Special Issues Guidelines
Editorial Process
Research and Publication Ethics
Article Processing Charges
Awards
Testimonials
Author Services
Initiatives
Sciforum
MDPI Books
Preprints.org
Scilit
SciProfiles
Encyclopedia
JAMS
Proceedings Series
About
Overview
Contact
Careers
News
Press
Blog
Sign In / Sign Up
Submit
Journals
Active Journals
Find a Journal
Proceedings Series
Topics
Information
For Authors
For Reviewers
For Editors
For Librarians
For Publishers
For Societies
For Conference Organizers
Open Access Policy
Institutional Open Access Program
Special Issues Guidelines
Editorial Process
Research and Publication Ethics
Article Processing Charges
Awards
Testimonials
Author Services
Initiatives
Sciforum
MDPI Books
Preprints.org
Scilit
SciProfiles
Encyclopedia
JAMS
Proceedings Series
About
Overview
Contact
Careers
News
Press
Blog
Sign In / Sign Up
Submit
 6.4
6.4

 4.5
Journals
Microorganisms
4.5
Journals
Microorganisms
 Rapid Isolation of Phages for the Treatment of Antibiotic Resistant Infections
Rapid Isolation of Phages for the Treatment of Antibiotic Resistant Infections
 Hospital-Acquired Pneumonia and Ventilator-Associated Pneumonia: A Literature Review
Hospital-Acquired Pneumonia and Ventilator-Associated Pneumonia: A Literature Review
 Streptococcus pyogenes, the Master of Enzymatic Immunomodulation
Streptococcus pyogenes, the Master of Enzymatic Immunomodulation
 Animal and In Vitro Models as Powerful Tools to Decipher the Effects of Enteric Pathogens on the Human Gut Microbiota
Animal and In Vitro Models as Powerful Tools to Decipher the Effects of Enteric Pathogens on the Human Gut Microbiota
 10-Year Experience of Pediatric Autoimmune Neuropsychiatric Disorders Associated with Streptococcal Infections (PANDAS)
Journal Description
Microorganisms
Microorganisms
is a scientific, peer-reviewed, open access journal of microbiology, published monthly online by MDPI. The Hellenic Society Mikrobiokosmos (MBK), the Spanish Society for Nitrogen Fixation (SEFIN) and the Society for Microbial Ecology and Disease (SOMED) are affiliated with the Microorganisms, and their members receive a discount on the article processing charges.
Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, PubAg, CAPlus / SciFinder, AGRIS, and other databases.
Journal Rank: JCR - Q2 (Microbiology) / CiteScore - Q2 (Microbiology (medical))
Rapid Publication: manuscripts are peer-reviewed and a first
decision is provided to authors approximately 15.1 days after submission; acceptance
to publication is undertaken in 2.8 days (median values for papers published in
this journal in the second half of 2023).
Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Testimonials: See what our editors and authors say about the Microorganisms.
Companion journal: Applied Microbiology.
Impact Factor:
4.5 (2022);
5-Year Impact Factor:
4.8 (2022)
subject
Imprint Information
get_app
Journal Flyer
Open Access
ISSN: 2076-2607
Latest Articles
attachment
Supplementary material:
Supplementary File 1 (ZIP, 102 KiB)
15 pages, 2949 KiB
Open AccessArticle
Microbiological Diversity and Associated Enzymatic Activities in Honey and Pollen from Stingless Bees from Northern Argentina
by
Virginia María Salomón, Johan Sebastian Hero, Andrés Hernán Morales, José Horacio Pisa, Luis María Maldonado, Nancy Vera, Rossana Elena Madrid and Cintia Mariana Romero
Microorganisms 2024, 12(4), 711; https://doi.org/10.3390/microorganisms12040711 (registering DOI) - 30 Mar 2024
Abstract
Honey and pollen from Tetragonisca fiebrigi and Scaptotrigona jujuyensis, stingless bees from northern Argentina, presented a particular microbiological profile and associated enzymatic activities. The cultured bacteria were mostly Bacillus spp. (44%) and Escherichia spp. (31%). The phylogenetic analysis showed a taxonomic distribution
[...] Read more.
Honey and pollen from Tetragonisca fiebrigi and Scaptotrigona jujuyensis, stingless bees from northern Argentina, presented a particular microbiological profile and associated enzymatic activities. The cultured bacteria were mostly Bacillus spp. (44%) and Escherichia spp. (31%). The phylogenetic analysis showed a taxonomic distribution according to the type of bee that was similar in both species. Microbial enzymatic activities were studied using hierarchical clustering. Bacillus spp. was the main bacterium responsible for enzyme production. Isolates with xylanolytic activity mostly presented cellulolytic activity and, in fewer cases, lipolytic activity. Amylolytic activity was associated with proteolytic activity. None of the isolated strains produced multiple hydrolytic enzymes in substantial amounts, and bacteria were classified according to their primary hydrolytic activity. These findings add to the limited knowledge of microbiological diversity in honey and pollen from stingless bees and also provide a physiological perspective of this community to assess its biotechnological potential in the food industry.
Full article
(This article belongs to the Topic Food Hygiene and Food Safety)
►▼
Show Figures
10-Year Experience of Pediatric Autoimmune Neuropsychiatric Disorders Associated with Streptococcal Infections (PANDAS)
Journal Description
Microorganisms
Microorganisms
is a scientific, peer-reviewed, open access journal of microbiology, published monthly online by MDPI. The Hellenic Society Mikrobiokosmos (MBK), the Spanish Society for Nitrogen Fixation (SEFIN) and the Society for Microbial Ecology and Disease (SOMED) are affiliated with the Microorganisms, and their members receive a discount on the article processing charges.
Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
High Visibility: indexed within Scopus, SCIE (Web of Science), PubMed, PMC, PubAg, CAPlus / SciFinder, AGRIS, and other databases.
Journal Rank: JCR - Q2 (Microbiology) / CiteScore - Q2 (Microbiology (medical))
Rapid Publication: manuscripts are peer-reviewed and a first
decision is provided to authors approximately 15.1 days after submission; acceptance
to publication is undertaken in 2.8 days (median values for papers published in
this journal in the second half of 2023).
Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Testimonials: See what our editors and authors say about the Microorganisms.
Companion journal: Applied Microbiology.
Impact Factor:
4.5 (2022);
5-Year Impact Factor:
4.8 (2022)
subject
Imprint Information
get_app
Journal Flyer
Open Access
ISSN: 2076-2607
Latest Articles
attachment
Supplementary material:
Supplementary File 1 (ZIP, 102 KiB)
15 pages, 2949 KiB
Open AccessArticle
Microbiological Diversity and Associated Enzymatic Activities in Honey and Pollen from Stingless Bees from Northern Argentina
by
Virginia María Salomón, Johan Sebastian Hero, Andrés Hernán Morales, José Horacio Pisa, Luis María Maldonado, Nancy Vera, Rossana Elena Madrid and Cintia Mariana Romero
Microorganisms 2024, 12(4), 711; https://doi.org/10.3390/microorganisms12040711 (registering DOI) - 30 Mar 2024
Abstract
Honey and pollen from Tetragonisca fiebrigi and Scaptotrigona jujuyensis, stingless bees from northern Argentina, presented a particular microbiological profile and associated enzymatic activities. The cultured bacteria were mostly Bacillus spp. (44%) and Escherichia spp. (31%). The phylogenetic analysis showed a taxonomic distribution
[...] Read more.
Honey and pollen from Tetragonisca fiebrigi and Scaptotrigona jujuyensis, stingless bees from northern Argentina, presented a particular microbiological profile and associated enzymatic activities. The cultured bacteria were mostly Bacillus spp. (44%) and Escherichia spp. (31%). The phylogenetic analysis showed a taxonomic distribution according to the type of bee that was similar in both species. Microbial enzymatic activities were studied using hierarchical clustering. Bacillus spp. was the main bacterium responsible for enzyme production. Isolates with xylanolytic activity mostly presented cellulolytic activity and, in fewer cases, lipolytic activity. Amylolytic activity was associated with proteolytic activity. None of the isolated strains produced multiple hydrolytic enzymes in substantial amounts, and bacteria were classified according to their primary hydrolytic activity. These findings add to the limited knowledge of microbiological diversity in honey and pollen from stingless bees and also provide a physiological perspective of this community to assess its biotechnological potential in the food industry.
Full article
(This article belongs to the Topic Food Hygiene and Food Safety)
►▼
Show Figures
 Figure 1 14 pages, 1054 KiB Open AccessArticle Molecular Identification and Subtype Analysis of Blastocystis sp. Isolates from Wild Mussels (Mytilus edulis) in Northern France by Manon Ryckman, Nausicaa Gantois, Ruben Garcia Dominguez, Jeremy Desramaut, Luen-Luen Li, Gaël Even, Christophe Audebert, Damien Paul Devos, Magali Chabé, Gabriela Certad, Sébastien Monchy and Eric Viscogliosi Microorganisms 2024, 12(4), 710; https://doi.org/10.3390/microorganisms12040710 (registering DOI) - 30 Mar 2024 Abstract Blastocystis sp. is the most common single-celled eukaryote colonizing the human gastrointestinal tract worldwide. Because of the proven zoonotic potential of this protozoan, sustained research is therefore focused on identifying various reservoirs of transmission to humans, and in particular animal sources. Numerous groups [...] Read more. Blastocystis sp. is the most common single-celled eukaryote colonizing the human gastrointestinal tract worldwide. Because of the proven zoonotic potential of this protozoan, sustained research is therefore focused on identifying various reservoirs of transmission to humans, and in particular animal sources. Numerous groups of animals are considered to be such reservoirs due to their handling or consumption. However, some of them, including mollusks, remain underexplored. Therefore, a molecular epidemiological survey conducted in wild mussels was carried out in Northern France (Hauts-de-France region) to evaluate the frequency and subtypes (STs) distribution of Blastocystis sp. in these bivalve mollusks. For this purpose, 100 mussels (Mytilus edulis) were randomly collected in two sampling sites (Wimereux and Dannes) located in the vicinity of Boulogne-sur-Mer. The gills and gastrointestinal tract of each mussel were screened for the presence of Blastocystis sp. by real-time polymerase chain reaction (qPCR) assay followed by direct sequencing of positive PCR products and subtyping through phylogenetic analysis. In parallel, sequences of potential representative Blastocystis sp. isolates that were previously obtained from temporal surveys of seawater samples at marine stations offshore of Wimereux were integrated in the present analysis. By taking into account the qPCR results from all mussels, the overall prevalence of the parasite was shown to reach 62.0%. In total, more than 55% of the positive samples presented mixed infections. In the remaining mussel samples with a single sequence, various STs including ST3, ST7, ST14, ST23, ST26 and ST44 were reported with varying frequencies. Such distribution of STs coupled with the absence of a predominant ST specific to these bivalves strongly suggested that mussels might not be natural hosts of Blastocystis sp. and might rather be carriers of parasite isolates from both human and animal (bovid and birds) waste. These data from mussels together with the molecular identification of isolates from marine stations were subsequently discussed along with the local geographical context in order to clarify the circulation of this protozoan in this area. The identification of human and animal STs of Blastocystis sp. in mussels emphasized the active circulation of this protozoan in mollusks and suggested a significant environmental contamination of fecal origin. This study has provided new insights into the host/carrier range and transmission of Blastocystis sp. and emphasized its potential as an effective sentinel species for water quality and environmental contamination. Full article (This article belongs to the Special Issue Seafood-Borne Pathogens) attachment Supplementary material: Supplementary File 1 (ZIP, 41922 KiB) 25 pages, 5825 KiB Open AccessArticle Analysis of Antimicrobial Resistance in Bacterial Pathogens Recovered from Food and Human Sources: Insights from 639,087 Bacterial Whole-Genome Sequences in the NCBI Pathogen Detection Database by Ashley L. Cooper, Alex Wong, Sandeep Tamber, Burton W. Blais and Catherine D. Carrillo Microorganisms 2024, 12(4), 709; https://doi.org/10.3390/microorganisms12040709 (registering DOI) - 30 Mar 2024 Abstract Understanding the role of foods in the emergence and spread of antimicrobial resistance necessitates the initial documentation of antibiotic resistance genes within bacterial species found in foods. Here, the NCBI Pathogen Detection database was used to query antimicrobial resistance gene prevalence in foodborne [...] Read more. Understanding the role of foods in the emergence and spread of antimicrobial resistance necessitates the initial documentation of antibiotic resistance genes within bacterial species found in foods. Here, the NCBI Pathogen Detection database was used to query antimicrobial resistance gene prevalence in foodborne and human clinical bacterial isolates. Of the 1,843,630 sequence entries, 639,087 (34.7%) were assigned to foodborne or human clinical sources with 147,788 (23.14%) from food and 427,614 (76.88%) from humans. The majority of foodborne isolates were either Salmonella (47.88%), Campylobacter (23.03%), Escherichia (11.79%), or Listeria (11.3%), and the remaining 6% belonged to 20 other genera. Most foodborne isolates were from meat/poultry (95,251 or 64.45%), followed by multi-product mixed food sources (29,892 or 20.23%) and fish/seafood (6503 or 4.4%); however, the most prominent isolation source varied depending on the genus/species. Resistance gene carriage also varied depending on isolation source and genus/species. Of note, Klebsiella pneumoniae and Enterobacter spp. carried larger proportions of the quinolone resistance gene qnrS and some clinically relevant beta-lactam resistance genes in comparison to Salmonella and Escherichia coli. The prevalence of mec in S. aureus did not significantly differ between meat/poultry and multi-product sources relative to clinical sources, whereas this resistance was rare in isolates from dairy sources. The proportion of biocide resistance in Bacillus and Escherichia was significantly higher in clinical isolates compared to many foodborne sources but significantly lower in clinical Listeria compared to foodborne Listeria. This work exposes the gaps in current publicly available sequence data repositories, which are largely composed of clinical isolates and are biased towards specific highly abundant pathogenic species. We also highlight the importance of requiring and curating metadata on sequence submission to not only ensure correct information and data interpretation but also foster efficient analysis, sharing, and collaboration. To effectively monitor resistance carriage in food production, additional work on sequencing and characterizing AMR carriage in common commensal foodborne bacteria is critical. Full article (This article belongs to the Special Issue Microbial Diversity and Antimicrobial Resistance Genes in the Environment) ►▼ Show Figures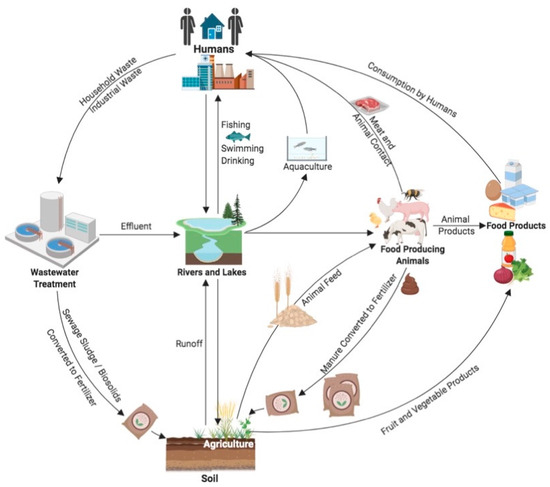 Figure 1 attachment Supplementary material: Supplementary File 1 (ZIP, 631 KiB) 17 pages, 8725 KiB Open AccessArticle The Vertical Metabolic Activity and Community Structure of Prokaryotes along Different Water Depths in the Kermadec and Diamantina Trenches by Hao Liu and Hongmei Jing Microorganisms 2024, 12(4), 708; https://doi.org/10.3390/microorganisms12040708 (registering DOI) - 30 Mar 2024 Abstract Prokaryotes play a key role in particulate organic matter’s decomposition and remineralization processes in the vertical scale of seawater, and prokaryotes contribute to more than 70% of the estimated remineralization. However, little is known about the microbial community and metabolic activity of the [...] Read more. Prokaryotes play a key role in particulate organic matter’s decomposition and remineralization processes in the vertical scale of seawater, and prokaryotes contribute to more than 70% of the estimated remineralization. However, little is known about the microbial community and metabolic activity of the vertical distribution in the trenches. The composition and distribution of prokaryotes in the water columns and benthic boundary layers of the Kermadec Trench and the Diamantina Trench were investigated using high-throughput sequencing and quantitative PCR, together with the Biolog EcoplateTM microplates culture to analyze the microbial metabolic activity. Microbial communities in both trenches were dominated by Nitrososphaera and Halobacteria in archaea, and by Alphaproteobacteria and Gammaproteobacteria in bacteria, and the microbial community structure was significantly different between the water column and the benthic boundary layer. At the surface water, amino acids and polymers were used preferentially; at the benthic boundary layers, amino acids and amines were used preferentially. Cooperative relationships among different microbial groups and their carbon utilization capabilities could help to make better use of various carbon sources along the water depths, reflected by the predominantly positive relationships based on the co-occurrence network analysis. In addition, the distinct microbial metabolic activity detected at 800 m, which was the lower boundary of the twilight zone, had the lowest salinity and might have had higher proportions of refractory carbon sources than the shallower water depths and benthic boundary layers. This study reflected the initial preference of the carbon source by the natural microbes in the vertical scale of different trenches and should be complemented with stable isotopic tracing experiments in future studies to enhance the understanding of the complex carbon utilization pathways along the vertical scale by prokaryotes among different trenches. Full article (This article belongs to the Special Issue Microbial Communities in Aquatic Environments) ►▼ Show Figures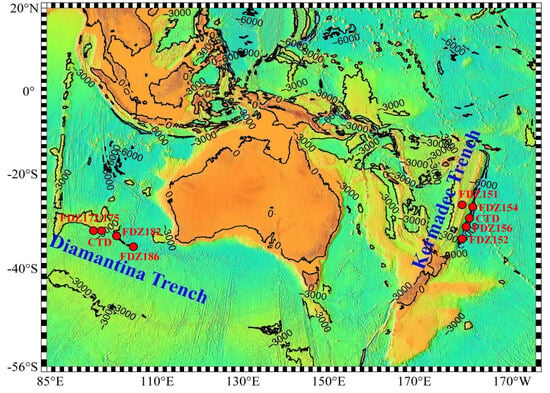 Figure 1 attachment Supplementary material: Supplementary File 1 (ZIP, 2421 KiB) 16 pages, 3957 KiB Open AccessArticle Expanded Archaeal Genomes Shed New Light on the Evolution of Isoprenoid Biosynthesis by Pengfei Zhu, Jialin Hou, Yixuan Xiong, Ruize Xie, Yinzhao Wang and Fengping Wang Microorganisms 2024, 12(4), 707; https://doi.org/10.3390/microorganisms12040707 (registering DOI) - 30 Mar 2024 Abstract Isoprenoids and their derivatives, essential for all cellular life on Earth, are particularly crucial in archaeal membrane lipids, suggesting that their biosynthesis pathways have ancient origins and play pivotal roles in the evolution of early life. Despite all eukaryotes, archaea, and a few [...] Read more. Isoprenoids and their derivatives, essential for all cellular life on Earth, are particularly crucial in archaeal membrane lipids, suggesting that their biosynthesis pathways have ancient origins and play pivotal roles in the evolution of early life. Despite all eukaryotes, archaea, and a few bacterial lineages being known to exclusively use the mevalonate (MVA) pathway to synthesize isoprenoids, the origin and evolutionary trajectory of the MVA pathway remain controversial. Here, we conducted a thorough comparison and phylogenetic analysis of key enzymes across the four types of MVA pathway, with the particular inclusion of metagenome assembled genomes (MAGs) from uncultivated archaea. Our findings support an archaeal origin of the MVA pathway, likely postdating the divergence of Bacteria and Archaea from the Last Universal Common Ancestor (LUCA), thus implying the LUCA’s enzymatic inability for isoprenoid biosynthesis. Notably, the Asgard archaea are implicated in playing central roles in the evolution of the MVA pathway, serving not only as putative ancestors of the eukaryote- and Thermoplasma-type routes, but also as crucial mediators in the gene transfer to eukaryotes, possibly during eukaryogenesis. Overall, this study advances our understanding of the origin and evolutionary history of the MVA pathway, providing unique insights into the lipid divide and the evolution of early life. Full article (This article belongs to the Section Microbiomes) ►▼ Show Figures Figure 1 25 pages, 4038 KiB Open AccessReview Biosynthesis Progress of High-Energy-Density Liquid Fuels Derived from Terpenes by Jiajia Liu, Man Lin, Penggang Han, Ge Yao and Hui Jiang Microorganisms 2024, 12(4), 706; https://doi.org/10.3390/microorganisms12040706 (registering DOI) - 30 Mar 2024 Abstract High-energy-density liquid fuels (HED fuels) are essential for volume-limited aerospace vehicles and could serve as energetic additives for conventional fuels. Terpene-derived HED biofuel is an important research field for green fuel synthesis. The direct extraction of terpenes from natural plants is environmentally unfriendly [...] Read more. High-energy-density liquid fuels (HED fuels) are essential for volume-limited aerospace vehicles and could serve as energetic additives for conventional fuels. Terpene-derived HED biofuel is an important research field for green fuel synthesis. The direct extraction of terpenes from natural plants is environmentally unfriendly and costly. Designing efficient synthetic pathways in microorganisms to achieve high yields of terpenes shows great potential for the application of terpene-derived fuels. This review provides an overview of the current research progress of terpene-derived HED fuels, surveying terpene fuel properties and the current status of biosynthesis. Additionally, we systematically summarize the engineering strategies for biosynthesizing terpenes, including mining and engineering terpene synthases, optimizing metabolic pathways and cell-level optimization, such as the subcellular localization of terpene synthesis and adaptive evolution. This article will be helpful in providing insight into better developing terpene-derived HED fuels. Full article (This article belongs to the Special Issue Yeasts Biochemistry and Biotechnology, 2nd Edition) ►▼ Show Figures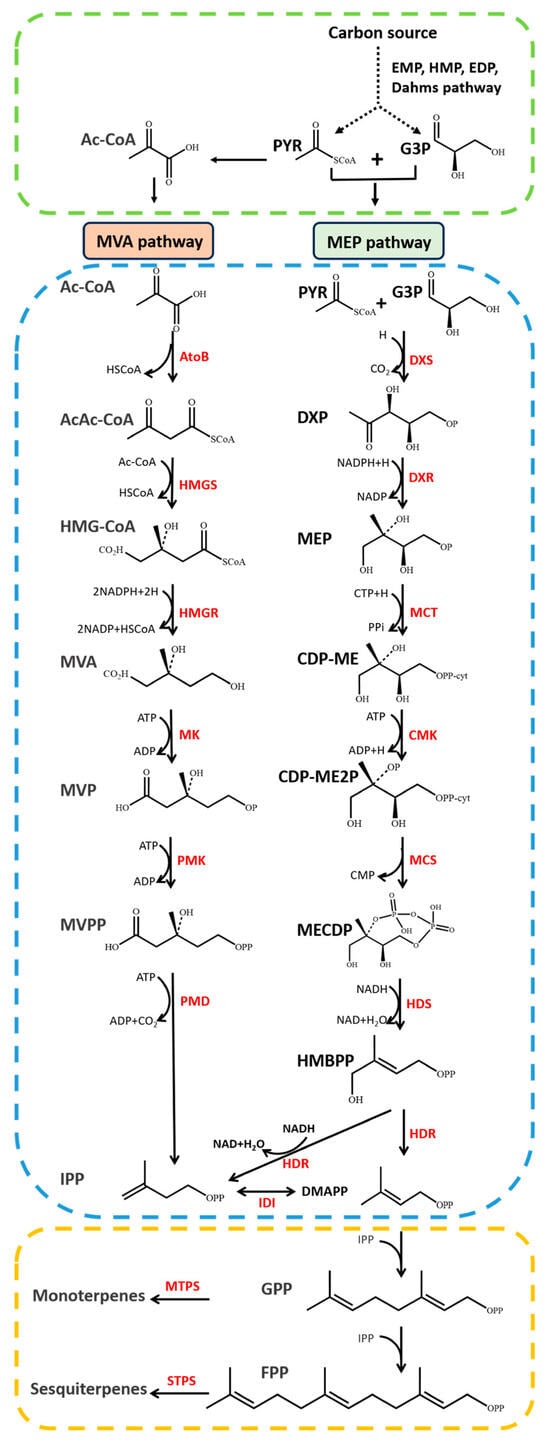 Figure 1 13 pages, 299 KiB Open AccessArticle Pseudomonas aeruginosa Bloodstream Infections Presenting with Septic Shock in Neutropenic Cancer Patients: Impact of Empirical Antibiotic Therapy by Cristina Royo-Cebrecos, Júlia Laporte-Amargós, Marta Peña, Isabel Ruiz-Camps, Carolina Garcia-Vidal, Edson Abdala, Chiara Oltolini, Murat Akova, Miguel Montejo, Malgorzata Mikulska, Pilar Martín-Dávila, Fabián Herrera, Oriol Gasch, Lubos Drgona, Hugo Manuel Paz Morales, Anne-Sophie Brunel, Estefanía García, Burcu Isler, Winfried V. Kern, Zaira R. Palacios-Baena, Guillermo Maestr de la Calle, Maria Milagro Montero, Souha S. Kanj, Oguz R. Sipahi, Sebnem Calik, Ignacio Márquez-Gómez, Jorge I. Marin, Marisa Z. R. Gomes, Philipp Hemmatii, Rafael Araos, Maddalena Peghin, Jose L. Del Pozo, Lucrecia Yáñez, Robert Tilley, Adriana Manzur, Andrés Novo, Jordi Carratalà and Carlota Gudioladd Show full author list remove Hide full author list Microorganisms 2024, 12(4), 705; https://doi.org/10.3390/microorganisms12040705 (registering DOI) - 30 Mar 2024 Abstract This large, multicenter, retrospective cohort study including onco-hematological neutropenic patients with Pseudomonas aeruginosa bloodstream infection (PABSI) found that among 1213 episodes, 411 (33%) presented with septic shock. The presence of solid tumors (33.3% vs. 20.2%, p < 0.001), a high-risk Multinational Association for [...] Read more. This large, multicenter, retrospective cohort study including onco-hematological neutropenic patients with Pseudomonas aeruginosa bloodstream infection (PABSI) found that among 1213 episodes, 411 (33%) presented with septic shock. The presence of solid tumors (33.3% vs. 20.2%, p < 0.001), a high-risk Multinational Association for Supportive Care in Cancer (MASCC) index score (92.6% vs. 57.4%; p < 0.001), pneumonia (38% vs. 19.2% p < 0.001), and infection due to multidrug-resistant P. aeruginosa (MDRPA) (33.8% vs. 21.1%, p < 0.001) were statistically significantly higher in patients with septic shock compared to those without. Patients with septic shock were more likely to receive inadequate empirical antibiotic therapy (IEAT) (21.7% vs. 16.2%, p = 0.020) and to present poorer outcomes, including a need for ICU admission (74% vs. 10.5%; p < 0.001), mechanical ventilation (49.1% vs. 5.6%; p < 0.001), and higher 7-day and 30-day case fatality rates (58.2% vs. 12%, p < 0.001, and 74% vs. 23.1%, p < 0.001, respectively). Risk factors for 30-day case fatality rate in patients with septic shock were orotracheal intubation, IEAT, infection due to MDRPA, and persistent PABSI. Therapy with granulocyte colony-stimulating factor and BSI from the urinary tract were associated with improved survival. Carbapenems were the most frequent IEAT in patients with septic shock, and the use of empirical combination therapy showed a tendency towards improved survival. Our findings emphasize the need for tailored management strategies in this high-risk population. Full article (This article belongs to the Special Issue Challenges in the Management of Infectious Complications in Neutropenic Cancer Patients) 21 pages, 3506 KiB Open AccessReview Elimination of Pathogen Biofilms via Postbiotics from Lactic Acid Bacteria: A Promising Method in Food and Biomedicine by Jiahao Che, Jingjing Shi, Chenguang Fang, Xiaoqun Zeng, Zhen Wu, Qiwei Du, Maolin Tu and Daodong Pan Microorganisms 2024, 12(4), 704; https://doi.org/10.3390/microorganisms12040704 (registering DOI) - 30 Mar 2024 Abstract Pathogenic biofilms provide a naturally favorable barrier for microbial growth and are closely related to the virulence of pathogens. Postbiotics from lactic acid bacteria (LAB) are secondary metabolites and cellular components obtained by inactivation of fermentation broth; they have a certain inhibitory effect [...] Read more. Pathogenic biofilms provide a naturally favorable barrier for microbial growth and are closely related to the virulence of pathogens. Postbiotics from lactic acid bacteria (LAB) are secondary metabolites and cellular components obtained by inactivation of fermentation broth; they have a certain inhibitory effect on all stages of pathogen biofilms. Postbiotics from LAB have drawn attention because of their high stability, safety dose parameters, and long storage period, which give them a broad application prospect in the fields of food and medicine. The mechanisms of eliminating pathogen biofilms via postbiotics from LAB mainly affect the surface adhesion, self-aggregation, virulence, and QS of pathogens influencing interspecific and intraspecific communication. However, there are some factors (preparation process and lack of target) which can limit the antibiofilm impact of postbiotics. Therefore, by using a delivery carrier and optimizing process parameters, the effect of interfering factors can be eliminated. This review summarizes the concept and characteristics of postbiotics from LAB, focusing on their preparation technology and antibiofilm effect, and the applications and limitations of postbiotics in food processing and clinical treatment are also discussed. Full article (This article belongs to the Special Issue New Strategies for Pathogenic Biofilms) ►▼ Show Figures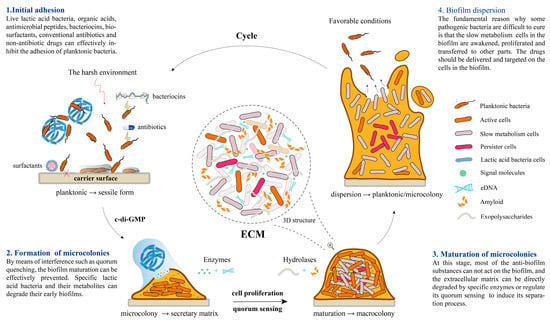 Figure 1 14 pages, 4592 KiB Open AccessArticle Impact of Organic Carbons Addition on the Enrichment Culture of Nitrifying Biofloc from Aquaculture Water: Process, Efficiency, and Microbial Community by Jiaqi Wu, Wujie Xu, Yu Xu, Haochang Su, Xiaojuan Hu, Yucheng Cao, Jianshe Zhang and Guoliang Wen Microorganisms 2024, 12(4), 703; https://doi.org/10.3390/microorganisms12040703 (registering DOI) - 30 Mar 2024 Abstract In this study, we developed a rapid and effective method for enriching the culture of nitrifying bioflocs (NBF) from aquacultural brackish water. The self-designed mixotrophic mediums with a single or mixed addition of sodium acetate, sodium citrate, and sucrose were used to investigate [...] Read more. In this study, we developed a rapid and effective method for enriching the culture of nitrifying bioflocs (NBF) from aquacultural brackish water. The self-designed mixotrophic mediums with a single or mixed addition of sodium acetate, sodium citrate, and sucrose were used to investigate the enrichment process and nitrification efficiency of NBF in small-scale reactors. The results showed that NBF with an MLVSSs from 1170.4 mg L−1 to 2588.0 mg L−1 were successfully enriched in a period of less than 16 days. The citrate group performed the fastest enrichment time of 10 days, while the sucrose group had the highest biomass of 2588.0 ± 384.7 mg L−1. In situ testing showed that the highest nitrification efficiency was achieved in the citrate group, with an ammonia oxidation rate of 1.45 ± 0.34 mg N L−1 h−1, a net nitrification rate of 2.02 ± 0.20 mg N L−1 h−1, and a specific nitrification rate of 0.72 ± 0.14 mg N g−1 h−1. Metagenomic sequencing revealed that Nitrosomonas (0.0~1.0%) and Nitrobacter (10.1~26.5%) were dominant genera for AOB and NOB, respectively, both of which had the highest relative abundances in the citrate group. Linear regression analysis further demonstrated significantly positive linear relations between nitrification efficiencies and nitrifying bacterial genera and gene abundance in NBF. The results of this study provide an efficient enrichment culture method of NBF for the operation of biofloc technology aquaculture systems, which will further promote its wide application in modern intensive aquaculture. Full article (This article belongs to the Special Issue Feature Papers in Microbial Biotechnology) ►▼ Show Figures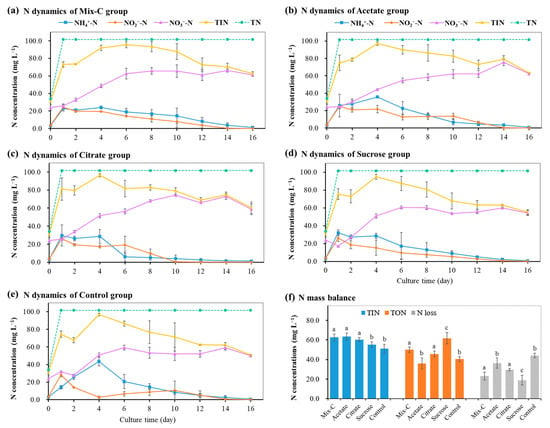 Figure 1 27 pages, 4646 KiB Open AccessReview Progress of Crude Oil Gasification Technology Assisted by Microorganisms in Reservoirs by Shumin Ni, Weifeng Lv, Zemin Ji, Kai Wang, Yuhao Mei and Yushu Li Microorganisms 2024, 12(4), 702; https://doi.org/10.3390/microorganisms12040702 (registering DOI) - 29 Mar 2024 Abstract Crude oil gasification bacteria, including fermenting bacteria, hydrocarbon-oxidizing bacteria, reducing bacteria, and methanogenic bacteria, participate in multi-step reactions involving initial activation, intermediate metabolism, and the methanogenesis of crude oil hydrocarbons. These bacteria degrade crude oil into smaller molecules such as hydrogen, carbon dioxide, [...] Read more. Crude oil gasification bacteria, including fermenting bacteria, hydrocarbon-oxidizing bacteria, reducing bacteria, and methanogenic bacteria, participate in multi-step reactions involving initial activation, intermediate metabolism, and the methanogenesis of crude oil hydrocarbons. These bacteria degrade crude oil into smaller molecules such as hydrogen, carbon dioxide, acetic acid, and formic acid. Ultimately, they convert it into methane, which can be utilized or stored as a strategic resource. However, the current challenges in crude oil gasification include long production cycles and low efficiency. This paper provides a summary of the microbial flora involved in crude oil gasification, the gasification metabolism pathways within reservoirs, and other relevant information. It specifically focuses on analyzing the factors that affect the efficiency of crude oil gasification metabolism and proposes suggestions for improving this efficiency. These studies deepen our understanding of the potential of reservoir ecosystems and provide valuable insights for future reservoir development and management. Full article (This article belongs to the Section Environmental Microbiology) ►▼ Show Figures Figure 1 17 pages, 8111 KiB Open AccessArticle Impacts of Arbuscular Mycorrhizal Fungi on Metabolites of an Invasive Weed Wedelia trilobata by Xinqi Jiang, Daiyi Chen, Yu Zhang, Misbah Naz, Zhicong Dai, Shanshan Qi and Daolin Du Microorganisms 2024, 12(4), 701; https://doi.org/10.3390/microorganisms12040701 (registering DOI) - 29 Mar 2024 Abstract The invasive plant Wedelia trilobata benefits in various aspects, such as nutrient absorption and environmental adaptability, by establishing a close symbiotic relationship with arbuscular mycorrhizal fungi (AMF). However, our understanding of whether AMF can benefit W. trilobata by influencing its metabolic profile remains [...] Read more. The invasive plant Wedelia trilobata benefits in various aspects, such as nutrient absorption and environmental adaptability, by establishing a close symbiotic relationship with arbuscular mycorrhizal fungi (AMF). However, our understanding of whether AMF can benefit W. trilobata by influencing its metabolic profile remains limited. In this study, Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was conducted to analyze the metabolites of W. trilobata under AMF inoculation. Metabolomic analysis identified 119 differentially expressed metabolites (DEMs) between the groups inoculated with AMF and those not inoculated with AMF. Compared to plants with no AMF inoculation, plants inoculated with AMF showed upregulation in the relative expression of 69 metabolites and downregulation in the relative expression of 50 metabolites. AMF significantly increased levels of various primary and secondary metabolites in plants, including amino acids, organic acids, plant hormones, flavonoids, and others, with amino acids being the most abundant among the identified substances. The identified DEMs mapped 53 metabolic pathways, with 7 pathways strongly influenced by AMF, particularly the phenylalanine metabolism pathway. Moreover, we also observed a high colonization level of AMF in the roots of W. trilobata, significantly promoting the shoot growth of this plant. These changes in metabolites and metabolic pathways significantly affect multiple physiological and biochemical processes in plants, such as free radical scavenging, osmotic regulation, cell structure stability, and material synthesis. In summary, AMF reprogrammed the metabolic pathways of W. trilobata, leading to changes in both primary and secondary metabolomes, thereby benefiting the growth of W. trilobata and enhancing its ability to respond to various biotic and abiotic stressors. These findings elucidate the molecular regulatory role of AMF in the invasive plant W. trilobata and provide new insights into the study of its competitive and stress resistance mechanisms. Full article (This article belongs to the Section Plant Microbe Interactions) ►▼ Show Figures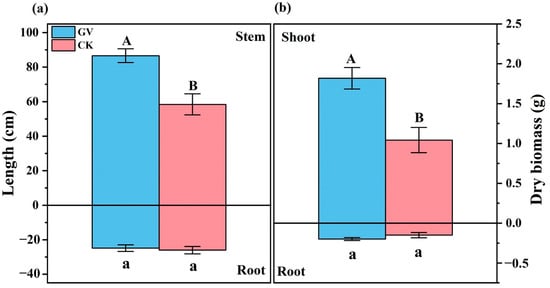 Figure 1 21 pages, 1910 KiB Open AccessReview Role of Plant-Growth-Promoting Rhizobacteria in Plant Machinery for Soil Heavy Metal Detoxification by Haichen Qin, Zixiao Wang, Wenya Sha, Shuhong Song, Fenju Qin and Wenchao Zhang Microorganisms 2024, 12(4), 700; https://doi.org/10.3390/microorganisms12040700 (registering DOI) - 29 Mar 2024 Abstract Heavy metals migrate easily and are difficult to degrade in the soil environment, which causes serious harm to the ecological environment and human health. Thus, soil heavy metal pollution has become one of the main environmental issues of global concern. Plant-growth-promoting rhizobacteria (PGPR) [...] Read more. Heavy metals migrate easily and are difficult to degrade in the soil environment, which causes serious harm to the ecological environment and human health. Thus, soil heavy metal pollution has become one of the main environmental issues of global concern. Plant-growth-promoting rhizobacteria (PGPR) is a kind of microorganism that grows around the rhizosphere and can promote plant growth and increase crop yield. PGPR can change the bioavailability of heavy metals in the rhizosphere microenvironment, increase heavy metal uptake by phytoremediation plants, and enhance the phytoremediation efficiency of heavy-metal-contaminated soils. In recent years, the number of studies on the phytoremediation efficiency of heavy-metal-contaminated soil enhanced by PGPR has increased rapidly. This paper systematically reviews the mechanisms of PGPR that promote plant growth (including nitrogen fixation, phosphorus solubilization, potassium solubilization, iron solubilization, and plant hormone secretion) and the mechanisms of PGPR that enhance plant–heavy metal interactions (including chelation, the induction of systemic resistance, and the improvement of bioavailability). Future research on PGPR should address the challenges in heavy metal removal by PGPR-assisted phytoremediation. Full article (This article belongs to the Special Issue Growth-Promoting Microorganisms as Potential Biological Control Agents of Plant Pests and Diseases) attachment Supplementary material: Supplementary File 1 (ZIP, 1145 KiB) 23 pages, 1340 KiB Open AccessArticle Pathogenomes of Shiga Toxin Positive and Negative Escherichia coli O157:H7 Strains TT12A and TT12B: Comprehensive Phylogenomic Analysis Using Closed Genomes by Anwar A. Kalalah, Sara S. K. Koenig, Peter Feng, Joseph M. Bosilevac, James L. Bono and Mark Eppinger Microorganisms 2024, 12(4), 699; https://doi.org/10.3390/microorganisms12040699 (registering DOI) - 29 Mar 2024 Abstract Shiga toxin-producing Escherichia coli are zoonotic pathogens that cause food-borne human disease. Among these, the O157:H7 serotype has evolved from an enteropathogenic O55:H7 ancestor through the displacement of the somatic gene cluster and recurrent toxigenic conversion by Shiga toxin-converting bacteriophages. However, atypical strains [...] Read more. Shiga toxin-producing Escherichia coli are zoonotic pathogens that cause food-borne human disease. Among these, the O157:H7 serotype has evolved from an enteropathogenic O55:H7 ancestor through the displacement of the somatic gene cluster and recurrent toxigenic conversion by Shiga toxin-converting bacteriophages. However, atypical strains that lack the Shiga toxin, the characteristic virulence hallmark, are circulating in this lineage. For this study, we analyzed the pathogenome and virulence inventories of the stx+ strain, TT12A, isolated from a patient with hemorrhagic colitis, and its respective co-isolated stx- strain, TT12B. Sequencing the genomes to closure proved critical to the cataloguing of subtle strain differentiating sequence and structural polymorphisms at a high-level of phylogenetic accuracy and resolution. Phylogenomic profiling revealed SNP and MLST profiles similar to the near clonal outbreak isolates. Their prophage inventories, however, were notably different. The attenuated atypical non-shigatoxigenic status of TT12B is explained by the absence of both the ΦStx1a- and ΦStx2a-prophages carried by TT12A, and we also recorded further alterations in the non-Stx prophage complement. Phenotypic characterization indicated that culture growth was directly impacted by the strains’ distinct lytic phage complement. Altogether, our phylogenomic and phenotypic analyses show that these intimately related isogenic strains are on divergent Stx(+/stx−) evolutionary paths. Full article (This article belongs to the Special Issue Microorganisms Associated with Infectious Disease 2.0) attachment Supplementary material: Supplementary File 1 (ZIP, 292 KiB) 17 pages, 916 KiB Open AccessArticle The Incidence and Risk Factors for Enterotoxigenic E. coli Diarrheal Disease in Children under Three Years Old in Lusaka, Zambia by Nsofwa Sukwa, Samuel Bosomprah, Paul Somwe, Monde Muyoyeta, Kapambwe Mwape, Kennedy Chibesa, Charlie Chaluma Luchen, Suwilanji Silwamba, Bavin Mulenga, Masiliso Munyinda, Seke Muzazu, Masuzyo Chirwa, Mwelwa Chibuye, Michelo Simuyandi, Roma Chilengi and Ann-Mari Svennerholm Microorganisms 2024, 12(4), 698; https://doi.org/10.3390/microorganisms12040698 - 29 Mar 2024 Abstract This study aimed to estimate the incidence and risk factors for Enterotoxigenic Escherichia coli (ETEC) diarrhea. This was a prospective cohort study of children recruited in a household census. Children were enrolled if they were 36 months or below. A total of 6828 [...] Read more. This study aimed to estimate the incidence and risk factors for Enterotoxigenic Escherichia coli (ETEC) diarrhea. This was a prospective cohort study of children recruited in a household census. Children were enrolled if they were 36 months or below. A total of 6828 children were followed up passively for 12 months to detect episodes of ETEC diarrhea. Diarrheal stool samples were tested for ETEC using colony polymerase chain reaction (cPCR). Among the 6828 eligible children enrolled, a total of 1110 presented with at least one episode of diarrhea. The overall incidence of ETEC diarrhea was estimated as 2.47 (95% confidence interval (CI): 2.10–2.92) episodes per 100 child years. Children who were HIV-positive (adjusted Hazard ratio (aHR) = 2.14, 95% CI: 1.14 to 3.99; p = 0.017) and those whose source of drinking water was public tap/borehole/well (aHR = 2.45, 95% CI: 1.48 to 4.06; p < 0.002) were at increased risk of ETEC diarrhea. This study found that children whose mothers have at least senior secondary school education (aHR = 0.49, 95% CI: 0.29 to 0.83; p = 0.008) were at decreased risk of ETEC diarrhea. Our study emphasizes the need for integrated public health strategies focusing on water supply improvement, healthcare for persons living with HIV, and maternal education. Full article (This article belongs to the Section Medical Microbiology) ►▼ Show Figures Figure 1 23 pages, 1640 KiB Open AccessReview Blue Biotechnology: Marine Bacteria Bioproducts by Karina Maldonado-Ruiz, Ruth Pedroza-Islas and Lorena Pedraza-Segura Microorganisms 2024, 12(4), 697; https://doi.org/10.3390/microorganisms12040697 - 29 Mar 2024 Abstract The ocean is the habitat of a great number of organisms with different characteristics. Compared to terrestrial microorganisms, marine microorganisms also represent a vast and largely unexplored reservoir of bioactive compounds with diverse industrial applications like terrestrial microorganisms. This review examines the properties [...] Read more. The ocean is the habitat of a great number of organisms with different characteristics. Compared to terrestrial microorganisms, marine microorganisms also represent a vast and largely unexplored reservoir of bioactive compounds with diverse industrial applications like terrestrial microorganisms. This review examines the properties and potential applications of products derived from marine microorganisms, including bacteriocins, enzymes, exopolysaccharides, and pigments, juxtaposing them in some cases against their terrestrial counterparts. We discuss the distinct characteristics that set marine-derived products apart, including enhanced stability and unique structural features such as the amount of uronic acid and sulfate groups in exopolysaccharides. Further, we explore the uses of these marine-derived compounds across various industries, ranging from food and pharmaceuticals to cosmetics and biotechnology. This review also presents a broad description of biotechnologically important compounds produced by bacteria isolated from marine environments, some of them with different qualities compared to their terrestrial counterparts. Full article (This article belongs to the Special Issue Bioactive Molecules from Microbial Sources) ►▼ Show Figures Graphical abstract attachment Supplementary material: Supplementary File 1 (ZIP, 312 KiB) 17 pages, 3527 KiB Open AccessArticle Comprehensive Genomics Investigation of Neboviruses Reveals Distinct Codon Usage Patterns and Host Specificity by Rahul Kaushik, Naveen Kumar, Pragya Yadav, Shubhankar Sircar, Anita Shete-Aich, Ankur Singh, Shailly Tomar, Thomas Launey and Yashpal Singh Malik Microorganisms 2024, 12(4), 696; https://doi.org/10.3390/microorganisms12040696 - 29 Mar 2024 Abstract Neboviruses (NeVs) from the Caliciviridae family have been linked to enteric diseases in bovines and have been detected worldwide. As viruses rely entirely on the cellular machinery of the host for replication, their ability to thrive in a specific host is greatly impacted [...] Read more. Neboviruses (NeVs) from the Caliciviridae family have been linked to enteric diseases in bovines and have been detected worldwide. As viruses rely entirely on the cellular machinery of the host for replication, their ability to thrive in a specific host is greatly impacted by the specific codon usage preferences. Here, we systematically analyzed the codon usage bias in NeVs to explore the genetic and evolutionary patterns. Relative Synonymous Codon Usage and Effective Number of Codon analyses indicated a marginally lower codon usage bias in NeVs, predominantly influenced by the nucleotide compositional constraints. Nonetheless, NeVs showed a higher codon usage bias for codons containing G/C at the third codon position. The neutrality plot analysis revealed natural selection as the primary factor that shaped the codon usage bias in both the VP1 (82%) and VP2 (57%) genes of NeVs. Furthermore, the NeVs showed a highly comparable codon usage pattern to bovines, as reflected through Codon Adaptation Index and Relative Codon Deoptimization Index analyses. Notably, yak NeVs showed considerably different nucleotide compositional constraints and mutational pressure compared to bovine NeVs, which appear to be predominantly host-driven. This study sheds light on the genetic mechanism driving NeVs’ adaptability, evolution, and fitness to their host species. Full article (This article belongs to the Special Issue Diversity and Pathogenesis of Common Human and Animal Viruses) ►▼ Show Figures Figure 1 17 pages, 5780 KiB Open AccessArticle Genomic and Phenotypic Analysis of Salmonella enterica Bacteriophages Identifies Two Novel Phage Species by Sudhakar Bhandare, Opeyemi U. Lawal, Anna Colavecchio, Brigitte Cadieux, Yella Zahirovich-Jovich, Zeyan Zhong, Elizabeth Tompkins, Margot Amitrano, Irena Kukavica-Ibrulj, Brian Boyle, Siyun Wang, Roger C. Levesque, Pascal Delaquis, Michelle Danyluk and Lawrence Goodridge Microorganisms 2024, 12(4), 695; https://doi.org/10.3390/microorganisms12040695 - 29 Mar 2024 Abstract Bacteriophages (phages) are potential alternatives to chemical antimicrobials against pathogens of public health significance. Understanding the diversity and host specificity of phages is important for developing effective phage biocontrol approaches. Here, we assessed the host range, morphology, and genetic diversity of eight Salmonella [...] Read more. Bacteriophages (phages) are potential alternatives to chemical antimicrobials against pathogens of public health significance. Understanding the diversity and host specificity of phages is important for developing effective phage biocontrol approaches. Here, we assessed the host range, morphology, and genetic diversity of eight Salmonella enterica phages isolated from a wastewater treatment plant. The host range analysis revealed that six out of eight phages lysed more than 81% of the 43 Salmonella enterica isolates tested. The genomic sequences of all phages were determined. Whole-genome sequencing (WGS) data revealed that phage genome sizes ranged from 41 to 114 kb, with GC contents between 39.9 and 50.0%. Two of the phages SB13 and SB28 represent new species, Epseptimavirus SB13 and genera Macdonaldcampvirus, respectively, as designated by the International Committee for the Taxonomy of Viruses (ICTV) using genome-based taxonomic classification. One phage (SB18) belonged to the Myoviridae morphotype while the remaining phages belonged to the Siphoviridae morphotype. The gene content analyses showed that none of the phages possessed virulence, toxin, antibiotic resistance, type I–VI toxin–antitoxin modules, or lysogeny genes. Three (SB3, SB15, and SB18) out of the eight phages possessed tailspike proteins. Whole-genome-based phylogeny of the eight phages with their 113 homologs revealed three clusters A, B, and C and seven subclusters (A1, A2, A3, B1, B2, C1, and C2). While cluster C1 phages were predominantly isolated from animal sources, cluster B contained phages from both wastewater and animal sources. The broad host range of these phages highlights their potential use for controlling the presence of S. enterica in foods. Full article (This article belongs to the Special Issue Bacteria Control by Phages) ►▼ Show Figures Figure 1 13 pages, 10309 KiB Open AccessArticle Pathogenicity of Aeromonas veronii Isolated from Diseased Macrobrachium rosenbergii and Host Immune-Related Gene Expression Profiles by Xiaojian Gao, Zhen Chen, Zirui Zhang, Qieqi Qian, Anting Chen, Lijie Qin, Xinzhe Tang, Qun Jiang and Xiaojun Zhang Microorganisms 2024, 12(4), 694; https://doi.org/10.3390/microorganisms12040694 - 29 Mar 2024 Abstract Aeromonas veronii is widespread in aquatic environments and is responsible for infecting various aquatic animals. In this study, a dominant strain was isolated from the hepatopancreas of diseased Macrobrachium rosenbergii and was named JDM1-1. According to its morphological, physiological, and biochemical characteristics and [...] Read more. Aeromonas veronii is widespread in aquatic environments and is responsible for infecting various aquatic animals. In this study, a dominant strain was isolated from the hepatopancreas of diseased Macrobrachium rosenbergii and was named JDM1-1. According to its morphological, physiological, and biochemical characteristics and molecular identification, isolate JDM1-1 was identified as A. veronii. The results of artificial challenge showed isolate JDM1-1 had high pathogenicity to M. rosenbergii with an LD50 value of 8.35 × 105 CFU/mL during the challenge test. Histopathological analysis revealed severe damage in the hepatopancreas and gills of the diseased prawns, characterized by the enlargement of the hepatic tubule lumen and gaps between the tubules as well as clubbing and degeneration observed at the distal end of the gill filament. Eight virulence-related genes, namely aer, ompA, lip, tapA, hlyA, flgA, flgM, and flgN, were screened by PCR assay. In addition, virulence factor detection showed that the JDM1-1 isolate produced lipase, lecithinase, gelatinase, and hemolysin. Furthermore, the mRNA expression profiles of immune-related genes of M. rosenbergii following A. veronii infection, including ALF1, ALF2, Crustin, C-lectin, and Lysozyme, were assessed, and the results revealed a significant upregulation in the hepatopancreas and intestines at different hours post infection. This study demonstrates that A. veronii is a causative agent associated with massive die-offs of M. rosenbergii and contributes valuable insights into the pathogenesis and host defense mechanisms of A. veronii invasion. Full article (This article belongs to the Special Issue Immunotoxic Factors Promoting Infectious Diseases in Cultured Marine Animals) ►▼ Show Figures Figure 1 14 pages, 1046 KiB Open AccessReview The Impact of Essential Amino Acids on the Gut Microbiota of Broiler Chickens by Thyneice Taylor-Bowden, Sarayu Bhogoju, Collins N. Khwatenge and Samuel N. Nahashon Microorganisms 2024, 12(4), 693; https://doi.org/10.3390/microorganisms12040693 - 29 Mar 2024 Abstract The research involving the beneficial aspects of amino acids being added to poultry feed pertaining to performance, growth, feed intake, and feed conversion ratio is extensive. Yet currently the effects of amino acids on the gut microbiota aren’t fully understood nor have there [...] Read more. The research involving the beneficial aspects of amino acids being added to poultry feed pertaining to performance, growth, feed intake, and feed conversion ratio is extensive. Yet currently the effects of amino acids on the gut microbiota aren’t fully understood nor have there been many studies executed in poultry to explain the relationship between amino acids and the gut microbiota. The overall outcome of health has been linked to bird gut health due to the functionality of gastrointestinal tract (GIT) for digestion/absorption of nutrients as well as immune response. These essential functions of the GI are greatly driven by the resident microbiota which produce metabolites such as butyrate, propionate, and acetate, providing the microbiota a suitable and thrive driven environment. Feed, age, the use of feed additives and pathogenic infections are the main factors that have an effect on the microbial community within the GIT. Changes in these factors may have potential effects on the gut microbiota in the chicken intestine which in turn may have an influence on health essentially affecting growth, feed intake, and feed conversion ratio. This review will highlight limited research studies that investigated the possible role of amino acids in the gut microbiota composition of poultry. Full article (This article belongs to the Special Issue Gastrointestinal Microbiome in Animals) ►▼ Show Figures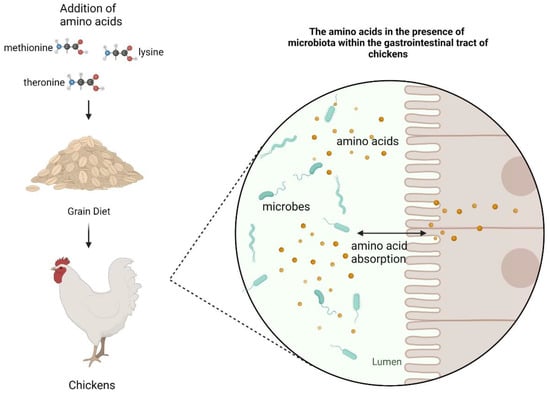 Figure 1 attachment Supplementary material: Supplementary File 1 (ZIP, 525 KiB) 17 pages, 13202 KiB Open AccessArticle The Bacterial and Fungal Compositions in the Rhizosphere of Asarum heterotropoides Fr. Schmidt var. mandshuricum (Maxim.) Kitag. in a Typical Planting Region by Fuqi Wang, Zilu Zhao, Yangyang Han, Shiying Li, Xinhua Bi, Shumeng Ren, Yingni Pan, Dongmei Wang and Xiaoqiu Liu Microorganisms 2024, 12(4), 692; https://doi.org/10.3390/microorganisms12040692 - 29 Mar 2024 Abstract Asarum is a traditional Chinese medicinal plant, and its dried roots are commonly used as medicinal materials. Research into the traits of the bacteria and fungus in the Asarum rhizosphere and how they relate to the potency of medicinal plants is important. During [...] Read more. Asarum is a traditional Chinese medicinal plant, and its dried roots are commonly used as medicinal materials. Research into the traits of the bacteria and fungus in the Asarum rhizosphere and how they relate to the potency of medicinal plants is important. During four cropping years and collecting months, we used ITS rRNA gene amplicon and sequencing to assess the population, diversity, and predominant kinds of bacteria and fungus in the rhizosphere of Asarum. HPLC was used to determine the three bioactive ingredients, namely asarinin, aristolochic acid I, and volatile oil. The mainly secondary metabolites of Asarum, relationships between microbial communities, soil physicochemical parameters, and possible influences on microbial communities owing to various cropping years and collecting months were all statistically examined. The cropping years and collecting months affected the abundance and diversity of rhizosphere bacteria and fungi, but the cropping year had a significant impact on the structures and compositions of the bacterial communities. The rhizosphere microorganisms were influenced by both the soil physicochemical properties and enzyme activities. Additionally, this study revealed that Trichoderma was positively correlated with the three bioactive ingredients of Asarum, while Tausonia showed entirely opposite results. Gibberella and Leptosphaeria demonstrated a significantly negative correlation with asarinin and violate oil, but they were weakly correlated with the aristolochic acid I content. This study revealed variations in the Asarum rhizosphere microorganism population, diversity, and dominant types across four cropping years and collecting months. The relationship between Asarum secondary metabolites, the soil physicochemical properties, enzyme activities, and rhizosphere microorganisms was discussed. Our results will guide the exploration of the soil characteristics and rhizosphere microorganisms’ structures by regulating the microbial community to enhance Asarum quality. Full article (This article belongs to the Special Issue Microbial Ecology of Soil Microorganisms, Networks with Plants and Ecosystem Services) ►▼ Show Figures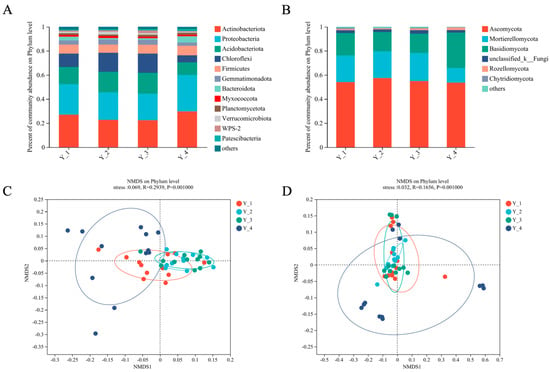 Figure 1 More Articles... Submit to Microorganisms
Review for Microorganisms
Share
Journal Menu
►
▼
Journal Menu
Microorganisms Home
Aims & Scope
Editorial Board
Reviewer Board
Topical Advisory Panel
Instructions for Authors
Special Issues
Topics
Sections & Collections
Article Processing Charge
Indexing & Archiving
Editor’s Choice Articles
Most Cited & Viewed
Journal Statistics
Journal History
Journal Awards
Society Collaborations
Editorial Office
Journal Browser
►
▼
Journal Browser
arrow_forward_ios
Forthcoming issue
arrow_forward_ios
Current issue
Vol. 12 (2024)
Vol. 11 (2023)
Vol. 10 (2022)
Vol. 9 (2021)
Vol. 8 (2020)
Vol. 7 (2019)
Vol. 6 (2018)
Vol. 5 (2017)
Vol. 4 (2016)
Vol. 3 (2015)
Vol. 2 (2014)
Vol. 1 (2013)
Highly Accessed Articles
View More...
Latest Books
Submit to Microorganisms
Review for Microorganisms
Share
Journal Menu
►
▼
Journal Menu
Microorganisms Home
Aims & Scope
Editorial Board
Reviewer Board
Topical Advisory Panel
Instructions for Authors
Special Issues
Topics
Sections & Collections
Article Processing Charge
Indexing & Archiving
Editor’s Choice Articles
Most Cited & Viewed
Journal Statistics
Journal History
Journal Awards
Society Collaborations
Editorial Office
Journal Browser
►
▼
Journal Browser
arrow_forward_ios
Forthcoming issue
arrow_forward_ios
Current issue
Vol. 12 (2024)
Vol. 11 (2023)
Vol. 10 (2022)
Vol. 9 (2021)
Vol. 8 (2020)
Vol. 7 (2019)
Vol. 6 (2018)
Vol. 5 (2017)
Vol. 4 (2016)
Vol. 3 (2015)
Vol. 2 (2014)
Vol. 1 (2013)
Highly Accessed Articles
View More...
Latest Books
 More Books and Reprints...
E-Mail Alert
News
4 March 2024
MDPI Insights: The CEO's Letter #9 - Romania, Research Integrity, Viruses
28 February 2024
Meet Us at the British Society for Parasitology Spring Meeting 2024, 2–5 April 2024, Liverpool, UK
More Books and Reprints...
E-Mail Alert
News
4 March 2024
MDPI Insights: The CEO's Letter #9 - Romania, Research Integrity, Viruses
28 February 2024
Meet Us at the British Society for Parasitology Spring Meeting 2024, 2–5 April 2024, Liverpool, UK
 9 February 2024
Recruiting Guest Editors for Microorganisms
9 February 2024
Recruiting Guest Editors for Microorganisms
 More News & Announcements...
Topics
Propose a Topic
Topic in
Biology, Diversity, Microorganisms, Molecules, Life
Extreme Environments: Microbial and Biochemical Diversity
Topic Editors: Massimiliano Fenice, Susanna Gorrasi, Marcella PasqualettiDeadline: 31 March 2024
Topic in
Biology, Ecologies, Forests, Microorganisms, Plants
Litter Decompositions: From Individuals to Ecosystems
Topic Editors: Wen Zhou, Guihua LiuDeadline: 30 April 2024
Topic in
Aquaculture Journal, Fishes, Microorganisms, Water
Women in Aquaculture Research
Topic Editors: Camino Ordás, Patrícia Díaz-RosalesDeadline: 30 June 2024
Topic in
Agronomy, Environments, Microorganisms, Pollutants, Sustainability, Water
Soil and Water Pollution Process and Remediation Technologies, 2nd Volume
Topic Editors: Hongbiao Cui, Ru Wang, Yu Shi, Haiying Lu, Lin ChenDeadline: 15 July 2024
More Topics
More News & Announcements...
Topics
Propose a Topic
Topic in
Biology, Diversity, Microorganisms, Molecules, Life
Extreme Environments: Microbial and Biochemical Diversity
Topic Editors: Massimiliano Fenice, Susanna Gorrasi, Marcella PasqualettiDeadline: 31 March 2024
Topic in
Biology, Ecologies, Forests, Microorganisms, Plants
Litter Decompositions: From Individuals to Ecosystems
Topic Editors: Wen Zhou, Guihua LiuDeadline: 30 April 2024
Topic in
Aquaculture Journal, Fishes, Microorganisms, Water
Women in Aquaculture Research
Topic Editors: Camino Ordás, Patrícia Díaz-RosalesDeadline: 30 June 2024
Topic in
Agronomy, Environments, Microorganisms, Pollutants, Sustainability, Water
Soil and Water Pollution Process and Remediation Technologies, 2nd Volume
Topic Editors: Hongbiao Cui, Ru Wang, Yu Shi, Haiying Lu, Lin ChenDeadline: 15 July 2024
More Topics
 Conferences
Announce Your Conference
More Conferences...
Special Issues
Propose a Special Issue
Special Issue in
Microorganisms
Vaginal Microbiome in Women's Health
Guest Editors: Sandra F. Borges, Sara Baptista da SilvaDeadline: 30 March 2024
Special Issue in
Microorganisms
Mobile Genetic Elements in Pathogens
Guest Editors: Maria Santagati, Francesco IannelliDeadline: 31 March 2024
Special Issue in
Microorganisms
Plant-Microbe Interaction State-of-the-Art Research in China
Guest Editors: Xianbing Wang, Jie Zhang, Wei Qian, Huiquan LiuDeadline: 15 April 2024
Special Issue in
Microorganisms
Plant and Human Probiotics: Consequences on the Autochthonous Microbiota
Guest Editors: Giorgia Novello, Francesco Vuolo, Elisa Gamalero, Elisa BonaDeadline: 30 April 2024
More Special Issues
Topical Collections
Topical Collection in
Microorganisms
Trends in Yeast Biochemistry and Biotechnology
Collection Editors: Seiji Shibasaki, Mitsuyoshi Ueda
Topical Collection in
Microorganisms
Epidemiology and Pathogenicity of Animal-Adapted Streptococci
Collection Editors: Peter Valentin-Weigand, Marcus Fulde
Topical Collection in
Microorganisms
Biodegradation and Environmental Microbiomes
Collection Editors: Shuangjiang Liu, Hongzhi Tang, Jiandong Jiang, Xiaolei Wu
Topical Collection in
Microorganisms
New Electrogenic Microbes
Collection Editor: Akihiro Okamoto
More Topical Collections
Microorganisms,
EISSN 2076-2607,
Published by MDPI
RSS
Content Alert
Further Information
Article Processing Charges
Pay an Invoice
Open Access Policy
Contact MDPI
Jobs at MDPI
Guidelines
For Authors
For Reviewers
For Editors
For Librarians
For Publishers
For Societies
For Conference Organizers
MDPI Initiatives
Sciforum
MDPI Books
Preprints.org
Scilit
SciProfiles
Encyclopedia
JAMS
Proceedings Series
Follow MDPI
LinkedIn
Facebook
Twitter
Conferences
Announce Your Conference
More Conferences...
Special Issues
Propose a Special Issue
Special Issue in
Microorganisms
Vaginal Microbiome in Women's Health
Guest Editors: Sandra F. Borges, Sara Baptista da SilvaDeadline: 30 March 2024
Special Issue in
Microorganisms
Mobile Genetic Elements in Pathogens
Guest Editors: Maria Santagati, Francesco IannelliDeadline: 31 March 2024
Special Issue in
Microorganisms
Plant-Microbe Interaction State-of-the-Art Research in China
Guest Editors: Xianbing Wang, Jie Zhang, Wei Qian, Huiquan LiuDeadline: 15 April 2024
Special Issue in
Microorganisms
Plant and Human Probiotics: Consequences on the Autochthonous Microbiota
Guest Editors: Giorgia Novello, Francesco Vuolo, Elisa Gamalero, Elisa BonaDeadline: 30 April 2024
More Special Issues
Topical Collections
Topical Collection in
Microorganisms
Trends in Yeast Biochemistry and Biotechnology
Collection Editors: Seiji Shibasaki, Mitsuyoshi Ueda
Topical Collection in
Microorganisms
Epidemiology and Pathogenicity of Animal-Adapted Streptococci
Collection Editors: Peter Valentin-Weigand, Marcus Fulde
Topical Collection in
Microorganisms
Biodegradation and Environmental Microbiomes
Collection Editors: Shuangjiang Liu, Hongzhi Tang, Jiandong Jiang, Xiaolei Wu
Topical Collection in
Microorganisms
New Electrogenic Microbes
Collection Editor: Akihiro Okamoto
More Topical Collections
Microorganisms,
EISSN 2076-2607,
Published by MDPI
RSS
Content Alert
Further Information
Article Processing Charges
Pay an Invoice
Open Access Policy
Contact MDPI
Jobs at MDPI
Guidelines
For Authors
For Reviewers
For Editors
For Librarians
For Publishers
For Societies
For Conference Organizers
MDPI Initiatives
Sciforum
MDPI Books
Preprints.org
Scilit
SciProfiles
Encyclopedia
JAMS
Proceedings Series
Follow MDPI
LinkedIn
Facebook
Twitter
 © 1996-2024 MDPI (Basel, Switzerland) unless otherwise stated
Disclaimer
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely
those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or
the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas,
methods, instructions or products referred to in the content.
Terms and Conditions
Privacy Policy
We use cookies on our website to ensure you get the best experience.
Read more about our cookies here.
Accept
Share Link
Copy
clear
Share
https://www.mdpi.com/journal/microorganisms
clear
Back to TopTop
© 1996-2024 MDPI (Basel, Switzerland) unless otherwise stated
Disclaimer
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely
those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or
the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas,
methods, instructions or products referred to in the content.
Terms and Conditions
Privacy Policy
We use cookies on our website to ensure you get the best experience.
Read more about our cookies here.
Accept
Share Link
Copy
clear
Share
https://www.mdpi.com/journal/microorganisms
clear
Back to TopTop
|
【本文地址】